Dogs’ noses decoded: Optical sensor unveils canine brain’s olfactory prowess
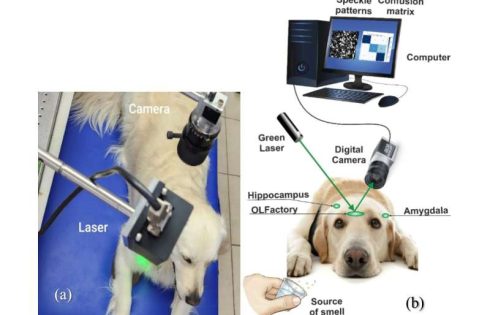
A pioneering study investigating the brain activity of dogs during scent detection has unveiled crucial insights into their remarkable olfactory capabilities. Researchers at Bar-Ilan University have developed an optical sensor capable of remote sensing dogs’ brain activity in three key regions—the olfactory bulb, hippocampus, and amygdala—that play a critical role in how dogs distinguish between different smells. This breakthrough could lead to the development of a compact, non-invasive device capable of interpreting and translating a dog’s olfactory perceptions for human understanding.
In the study published in the Journal of Biophotonics, scientists employed a cutting-edge detection structure system using laser technology and a high-resolution camera to capture brain activity in real-time from four dog breeds.
These dogs were exposed to four distinct scent stimuli—garlic, menthol, alcohol, and marijuana. The data were then analyzed using a machine-learning algorithm revealing that the amygdala plays a significant role in scent differentiation, highlighting the emotional and memory-related aspects of odor processing.
“The findings show that the amygdala is crucial in the way dogs process and react to odors, with specific scents triggering distinct emotional and memory responses, and we are capable of optically detecting their brain activity in this region,” said Prof. Zeev Zalevsky, from the Kofkin Faculty of Engineering at Bar-Ilan University. “This discovery could be the first step toward creating a device that enables us to better understand and interpret the unique way dogs perceive and differentiate smells.”
The study introduces an innovative method of brain activity analysis through laser-based speckle pattern detection, a remote, non-invasive technique that has never been applied to canine brain activity. Unlike traditional methods such as fMRI or EEG, this approach allows researchers to observe brain responses without requiring the dog to be sedated or confined to bulky equipment. This opens up new possibilities for studying dogs in real-world environments, making the technique both affordable and accessible for further research.
Dogs have long been celebrated for their exceptional sense of smell, and this research further illuminates the advanced processes that occur in their brains when detecting odors. With an olfactory system far more developed than humans, dogs can detect a broader range of odors, with specialized receptors in their noses that allow them to process and distinguish even the faintest scents.
This new research offers a glimpse into the intricate workings of the canine brain as it processes different smells, presenting a promising avenue for future applications in areas such as drug detection, medical diagnostics, and search-and-rescue missions.
“Our next step is to develop a portable, Wi-Fi-controlled device equipped with a mini camera and laser system, which could be mounted on a dog’s head and used to monitor its olfactory responses in real time,” said Dr. Yafim Beiderman from Prof. Zalevsky’s Optical Research Lab at Bar-Ilan University.
“This could significantly enhance the way dogs are used in scent detection, from detecting illegal substances to diagnosing diseases in humans, all while deepening our understanding of how they perceive the world around them. More importantly, this real-time sensing could bypass the need to train dogs to utilize their scent abilities.”
The implications of this research could also revolutionize the way dogs are utilized in law enforcement, health care, and beyond. As dogs continue to be invaluable partners in scent detection, this device could provide a means of translating their highly specialized abilities into data that is useful for humans, fostering a stronger connection between the two species.