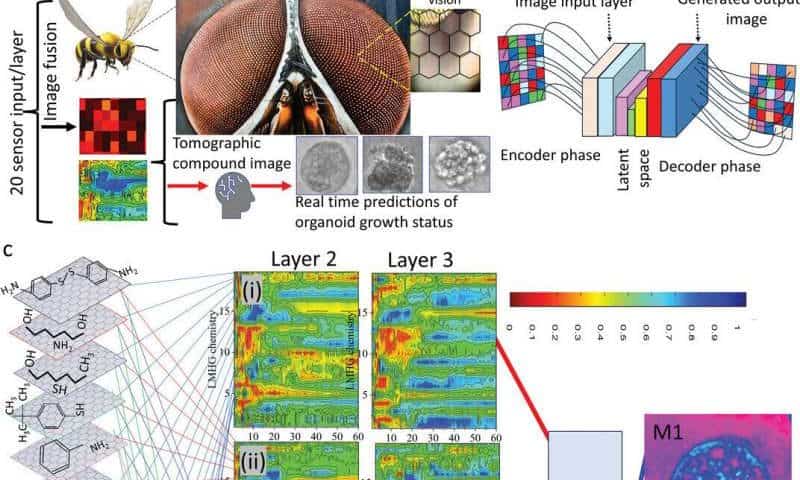
Insect vision inspires noninvasive method for deep tissue molecular mapping
The journal Advanced Materials recently published a study introducing a new method for monitoring molecular processes deep within tissue. Developed at the Technion–Israel Institute of Technology, the innovation is expected to accelerate key advancements in personalized medicine, cancer diagnosis, and early disease detection.
The research was led by Prof. Hossam Haick, postdoctoral fellow Dr. Arnab Maity, and Ph.D. student Vivian Darsa Maidantchik from the Wolfson Faculty of Chemical Engineering at the Technion. The study also involved Dr. Dalit Barkan, research assistant Dr. Keren Weidenfeld, and Prof. Sarit Larisch from the University of Haifa’s Faculty of Natural Sciences.
The Technion researchers’ method enables functional and molecular mapping of organoids—three-dimensional cell-based models that replicate the structural and functional characteristics of natural tissues. Organoids play a critical role in biomedical research by allowing scientists to study health and disease states and assess the effects of various treatments on organs and tissues.
Despite their potential, organoids face major technological challenges, particularly in monitoring internal tissue processes. Current methods are expensive and have the following significant limitations:
- Some techniques destroy the tissue (e.g., RNA sequencing)
- Others are blind to deep-tissue processes (e.g., confocal microscopy)
Technion’s breakthrough overcomes these limitations with a low-cost, accurate, and non-invasive approach that allows for continuous monitoring of structural and molecular changes within organoids.
Chemical tomography: New method in deep-tissue monitoring
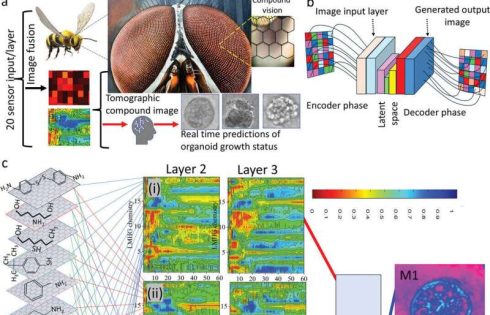
The researchers’ new method, called chemical tomography, provides insights into tissue function by analyzing volatile organic compounds (VOCs). These molecules are present in exhaled breath, saliva, sweat, and other bodily fluids. Prof. Haick is a leading global expert in the use of VOCs for early disease detection. His prior research has led to the development of multiple diagnostic technologies based on VOC analysis.
In this study, VOC monitoring enabled the dynamic molecular and functional mapping of a human breast tissue organoid, revealing key protein and genomic data associated with the transformation of healthy breast tissue into cancerous tissue.
The system detects VOCs using a graphene-based sensor array, with the collected data analyzed through generative artificial intelligence (AI). The inspiration for this technology comes from the compound eye of insects—a structure composed of multiple small eyes that send numerous images to the insect’s brain for analysis. In the system, the graphene sensors function as the compound eye, while AI acts as the brain, processing and interpreting the data.
The new system provides real-time, dynamic mapping of organoids at a significantly lower cost than existing alternatives, without damaging the tested tissue. This method enables researchers to:
- Track cancer progression at different stages
- Gain a deeper understanding of cancer biology
- Map biochemical pathways, metabolic markers, and molecular processes involved in cancer development
Using this new approach, the researchers identified six biochemical pathways responsible for producing 12 different types of VOCs, which could serve as biomarkers for disease states.
According to Prof. Haick, “Beyond cancer applications, our system has the potential to diagnose issues in various organs, including the kidneys, brain, and liver. It could also transmit real-time internal health data to an external monitoring system via an antenna, enabling continuous tracking of tissue health and early disease warnings. This is a breakthrough in integrating artificial intelligence into medicine, particularly in personalized health care.”