Northwestern University researchers have identified structural features in engineered cell receptors that correlate with variations in receptor function.
Computational protein structure prediction tools were used to analyze a library of synthetic receptors, revealing that specific structural attributes such as ectodomain (ECD) distance and transmembrane domain (TMD) interactions are associated with receptor performance.
Engineered cell therapies rely on synthetic receptors to transduce external signals into intracellular responses. The precise relationship between receptor structure and function remains poorly understood. Advances in protein structure prediction tools, such as AlphaFold and ColabFold, have enabled the modeling of complex proteins, including single-pass transmembrane receptors.
In the study, “Exploring Structure-Function Relationships in Engineered Receptor Performance Using Computational Structure Prediction,” published in GEN Biotechnology, researchers integrated predicted protein conformations with experimental performance metrics across four engineered receptor families, examining whether certain molecular features correlated with functional output.
Researchers analyzed a library of engineered receptors derived from natural cytokine receptors, focusing on Modular Extracellular Sensor Architecture (MESA) receptors. MESA receptors use ligand-induced dimerization to activate a split Tobacco Etch Virus Protease (TEVp), which releases a transcription factor to drive gene expression.
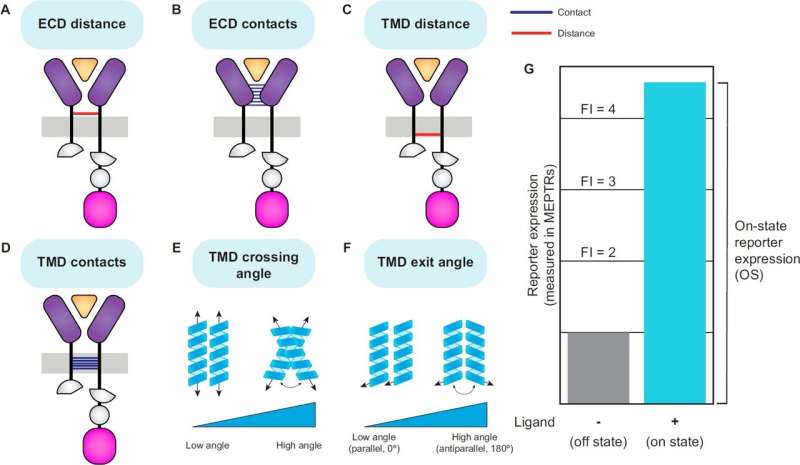
ColabFold was used to generate structural models of receptor ECDs in complex with their ligands, while PREDDIMER was employed to model TMD interactions.
Structural features were quantified, including ECD distance, ECD contacts, TMD distance, TMD contacts, TMD crossing angle, and TMD exit angle. Receptor performance was assessed based on on-state reporter expression (OS) and fold induction (FI) in experimental data.
Substantial variation in receptor performance was explained by structural features. Shorter ECD and TMD distances correlated with higher OS, while greater ECD and TMD contacts were also associated with improved performance. TMD exit angle had a significant relationship with receptor function in certain receptor families, particularly in tumor necrosis factor (TNF) MESA.
Structural models explain less variation in performance for heterotetrameric receptors, such as interleukin-10 (IL-10) MESA and transforming growth factor beta (TGFb) MESA. This suggests that additional factors, such as higher-order receptor complex formation, may contribute more to the performance of these receptors.
To complement structural analyses, the researchers evaluated categorical variables representing domain choices within receptor chains. One-hot encoding of receptor components revealed that VEGF MESA receptors containing a CD28 TMD in the C-terminal chain and a VEGFR1 ECD exhibited higher OS. The reduced categorical model explained 57% of OS variation, closely aligning with the structural model’s performance.
A combined model integrating both structural and categorical features demonstrated superior predictive power, explaining 79% of OS variation. This improvement indicates that while structural and categorical models share some information, they capture distinct determinants of receptor performance.
The findings present actionable hypotheses for synthetic receptor engineering. For instance, modifying receptor linkers to enhance ECD contacts or selecting TMD pairs that favor closer C-terminal proximities could improve receptor function.
A notable challenge in this work was modeling the entire single-pass transmembrane receptor,including the natural ectodomain (ECD), the transmembrane domain (TMD), and the split TEVp intracellular domains (NTEVp or CTEVp) using ColabFold.
Specifically, the tool struggled with aligning the shorter TEVp regions within long receptor sequences. During the multiple sequence alignment (MSA) search step, the sizable ECD and native TMD often dominated the alignment results, causing ColabFold to drop or poorly cover the TEVp segments.
As a result, the predicted structures sometimes failed to show a properly folded NTEVp or CTEVp, yielding low-confidence scores (pLDDT) in those intracellular portions.
Shorter ectodomains (e.g., IL-10 receptors) were more likely to retain coverage for the TEVp domains, allowing a few full-length models to converge on structures where the NTEVp and CTEVp were reconstituted.
Because of these alignment issues, the study opted for a modular approach, modeling ECD–ligand complexes separately with ColabFold and then using PREDDIMER to capture TMD–TMD interactions.
While this strategy prevented direct visualization of how NTEVp and CTEVp might orient themselves under membrane constraints, it ensured higher confidence in the predicted extracellular and transmembrane regions. The trade-off is that intermolecular effects were not fully captured.