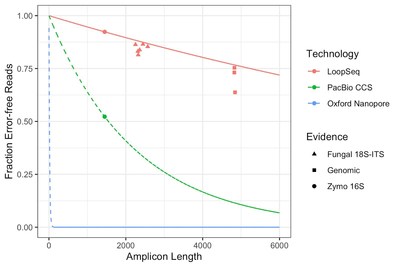
Loop Genomics long read shown to have an accuracy of 99.995% out to 6kb
SAN JOSE, Calif. — A new pre-print runs a head-to-head comparison between Loop Genomics, PacBio and Oxford Nanopore’s accuracy in long read microbiome sequencing.
In this new preprint, Callahan et al, the creators of DADA2, record LoopSeq’s long-read microbiome sequencing’s accuracy at 99.995% up to 6kb using Illumina sequencers, well above PacBio’s accuracy and orders of magnitude above Oxford Nanopore’s.
“We are pleased to see this external validation of Loop’s long-read accuracy in a domain as important as the microbiome. Labs have been looking to start using long reads but have been waiting for long read technology to improve. Long reads are the future of sequencing but we won’t get there before long-reads are accurate, trustworthy and widely accessible. That is our mission,” says Tuval Ben Yehezkel PhD, CEO and founder of Loop Genomics.
Leveraging the same accurate long-read technology described in the pre-print Loop Genomics is using Illumina sequencers to deliver long-read data in Immune-repertoire sequencing, CRISPR editing validation, Viral, Phage and microbiome sequencing. Most recently, Loops launched a massively parallel solution for sequencing isolates in large scale Synthetic Biology projects.
“Our goal is to enable researchers to gain valuable insight that only highly accurate long-read sequencing data can provide them using a standard Illumina sequencer and Loop Genomics’ services or kits,” says Tuval Ben Yehezkel PhD, CEO and founder of Loop Genomics.
Founded in late 2015, Loop Genomics is a San Francisco bay area company that commercializes technology for long-read sequencing using Illumina sequencers. The company offers various service and kits for long-read sequencing using Illumina sequencers.