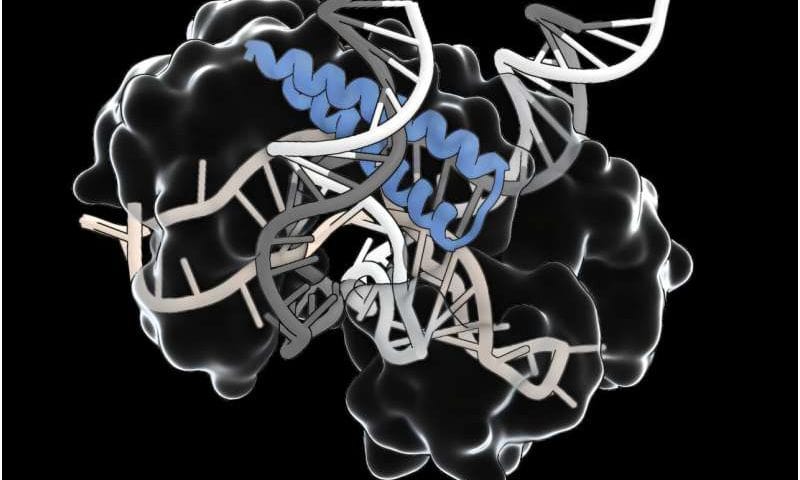
A vast search of natural diversity has led scientists at MIT’s McGovern Institute and the Broad Institute of MIT and Harvard to uncover ancient systems with the potential to expand the genome-editing toolbox. These systems, which the researchers call TIGR (Tandem Interspaced Guide RNA) systems, use RNA to guide them to specific sites on DNA.
TIGR systems can be reprogrammed to target any DNA sequence of interest, and they have distinct functional modules that can act on the targeted DNA. In addition to its modularity, TIGR is very compact compared to other RNA-guided systems, like CRISPR, which is a major advantage for delivering it in a therapeutic context.
These findings appear in the journal Science.
“This is a very versatile RNA-guided system with a lot of diverse functionalities,” says Feng Zhang, the James and Patricia Poitras Professor of Neuroscience at MIT who led the research. The TIGR-associated (Tas) proteins that Zhang’s team found share a characteristic RNA-binding component that interacts with an RNA guide that directs it to a specific site in the genome. Some cut the DNA at that site, using an adjacent DNA-cutting segment of the protein. That modularity could facilitate tool development, allowing researchers to swap useful new features into natural Tas proteins.
“Nature is pretty incredible,” remarks Zhang, who is also an investigator at the McGovern Institute and the Howard Hughes Medical Institute, a core member of the Broad Institute, a professor of brain and cognitive sciences and biological engineering at MIT, and co-director of the K. Lisa Yang and Hock E. Tan Center for Molecular Therapeutics at MIT.
“It’s got a tremendous amount of diversity, and we have been exploring that natural diversity to find new biological mechanisms and harnessing them for different applications to manipulate biological processes,” he says.
Previously, Zhang’s team had adapted bacterial CRISPR systems into gene-editing tools that have transformed modern biology. His team has also found a variety of programmable proteins, both from CRISPR systems and beyond.
In their new work, to find novel programmable systems, the team began by zeroing in on a structural feature of the CRISPR Cas9 protein that binds to the enzyme’s RNA guide. That is a key feature that has made Cas9 such a powerful tool.
“Being RNA-guided makes it relatively easy to reprogram, because we know how RNA binds to other DNA or other RNA,” Zhang explains. His team searched hundreds of millions of biological proteins with known or predicted structures, looking for any that shared a similar domain. To find more distantly related proteins, they used an iterative process: from Cas9, they identified a protein called IS110, which had previously been shown by others to bind RNA. They then zeroed in on the structural features of IS110 that enable RNA binding and repeated their search.
At this point, the search had turned up so many distantly related proteins that the team turned to artificial intelligence to make sense of the list.
“When you are doing iterative, deep mining, the resulting hits can be so diverse that they are difficult to analyze using standard phylogenetic methods, which rely on conserved sequences,” explains Guilhem Faure, a computational biologist in Zhang’s lab.